- 07 December 2021
The CS3MESH4EOSC Project is developing several application-specific extensions to be integrated with the protocols and application programming interfaces (APIs) developed for the Science Mesh. With them, the different applications can be fully integrated into the Science Mesh and, therefore, will be available to the Science Mesh.
The Science Mesh is an interoperable platform, which aims to make it easy for users to sync and share data across institutional and national boundaries. By joining the Science Mesh, these services will extend their functionalities, while having access to a wide community of potential end-users that aim to assemble their data in an efficient, reliable, collaborative, interoperable, and transparent way
Customised Extensions to Serve Different Data Management Needs
The Science Mesh has 4 main categories of data services, which are being populated by CS3MESH4EOSC through the integration of different existing applications developed by third-party initiatives:
- Data Science Environments: enable sharing of computational tools, algorithms, and resources across the Science Mesh;
- Open Data Systems: handle structured research-related data and use it for activities typically associated with it (archive, deposit and publish);
- Collaborative Documents: cross-federation collaborative content-editing applications which allow users to work together in real time;
- On-demand Data Transfers: High-speed transfer of information from remote locations to local sites across different countries, specifically supporting use-cases where it is not possible to extend processing capabilities to remote sites.
With regards to Data Science Environments, one of the applications being integrated into the Science Mesh is JupyterLab - a web-based interactive development environment for Jupyter notebooks and associated data. Its integration into the Science Mesh led to the development of a dedicated extension called cs3api4lab, which provides it with file browsing and additional share and collaboration functionalities for notebooks and resources in a distributed setting. The back-end part of the JupyterLab extension is implemented by replacing the Content Manager and Checkpoints components of the JupyterLab, as well as providing Representational State Transfer (REST) endpoints for integration with the front-end.

Figure: JupyterLab – CS3 API integration back-end component architecture
On the subject of Open Data Systems, Describo is a web-based tool that allows end users to define data collections, annotate them and render them into standardised packages. The key enabler for this is the Research Object Crate (RO-Crate) standard, which has been selected as the packaging and description data format for Describo and the Science Mesh. Describo will allow Science Mesh users to enrich their datasets with structured metadata, so that they can later be transferred to archival and repository systems. The integration of Describo into the Science Mesh is made possible by Reva (and the CS3APIS), which allow for seamless interaction with backend storage systems.
In order to select the RO-Crates, the user will use a web-based tool called ScieboRDS. The tool also allows this service offer to other third-party systems (e.g. archives, repositories and publishers). A practical example of how these two applications can be used together is starting with ScieboRDS (e.g., “a scientist wants to publish a paper with a certain dataset attached”), hand it over to the Describo environment (“select the relevant data to be crated up”) and then hand back to ScieboRDS (“here’s the crated and metadata-augmented data the scientist wanted to publish”).
Regarding Collaborative Documents, Collabora Online, OnlyOffice and CodiMD all now have proof-of-concept implementations running on the mesh. Collabora Online is a powerful open-source software (OSS) online office suite that supports all major document, spreadsheet and presentation file formats, with good collaborative editing and office file format support. OnlyOffice is another OSS office suite that features online document editors, platform for document management, corporate communication, mail, and project management tools. CodiMD is also OSS and offers a, web-based, self-hosted, collaborative markdown editor, used to collaborate on notes, graphs and presentations in real-time.
Last but not least, the On-demand Data Transfers service is relying on Rclone - a tool that is able to synchronise and copy data to and from cloud storage services. It’s binding into the Science Mesh is based on Open Cloud Mesh (OCM) and CS3 APIs, to allow that a user from a specific EFSS can transfer data directly to a user in another EFSS. The FTS, a batch scheduler of data transfers, is also used in the situations where transfers are initiated by EFSS service but need offloaded to a secondary service (the FTS) to better orchestrate data transfers around the globe. FTS can also be used in conjunction with RUCIO, a scientific data management system used as well to organise petabytes of data located in heterogeneous storage. The Rucio/FTS-based workflow will be developed in the coming months.
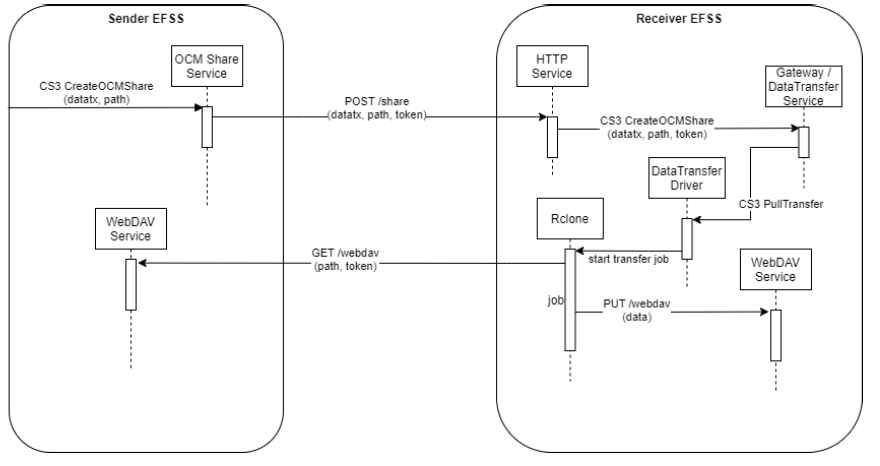
Figure 2: Direct data transfer; pull model work-flow
What Lies Ahead?
The Science Mesh is a dynamic platform where technologies developed by third-party organisations can be integrated it, to easily sync and share data. While there are already prototypes for a series of services and applications which integrate with the Mesh, its very design philosophy is founded on the possibility to enrich it further with new tools and workflows. To that end, new integration components and extensions to the various APIs and protocols will surely be developed along the way. This work will be done, whenever possible, in partnership with the initiatives behind those same technologies and capitalising on existing proven standards.




















